LARGE DISCREPANCY IN PNEUMOCOCCAL CARRIAGE RATE DEPENDING ON SITE OF SAMPLING AND METHOD OF DETECTION (ID 306)
- Michaela Tinggaard, Denmark
- Michaela Tinggaard, Denmark
- Hans-Christian Slotved, Denmark
- Nichlas Hovmand, Denmark
- Thomas Benfield, Denmark
A NEW CONVENTIONAL-PCR STRATEGY FOR DIFFERENTIAL DETECTION OF S.PNEUMONIAE AND S.PSEUDOPNEUMONIAE FROM NON-STERILE-SOURCES CONFIRMS HIGH LEVELS OF NASOPHARYNGEAL CARRIAGE AMONG HEALTHY OLDER-ADULTS FROM MEDELLÍN, COLOMBIA (ID 398)
- Santiago Cardona, Colombia
- Santiago Cardona, Colombia
- Juliana Andrea Carmona, Colombia
- Laura Leandra Beltrán, Colombia
- Andrés Felipe Castro, Colombia
- María Alejandra Palacio, Colombia
- José Luis Torres, Colombia
- Johan Alexis Bolivar, Colombia
- Leidy Johana Acevedo, Colombia
- Laura Mery Muñoz, Colombia
- Luis Felipe Vélez, Colombia
- Yulieth Alexandra Zúñiga, Colombia
- Anlly Banessa Holguín, Colombia
- Miriam Estela Molina, Colombia
- John Jairo Ruiz, Colombia
- Deissy Vargas, Colombia
- Catalina María Arboleda, Colombia
- Jessica Lorena Morales, Colombia
- Alejandro Gómez, Germany
- Sven Hammerschmidt, Germany
- Gustavo Adolfo Gámez, Colombia
A NEW FLEXIBLE-GENOMIC TWO-COMPONENT REGULATORY SYSTEM FOR STREPTOCOCCUS PNEUMONIAE AND THE IMPACT OF PNEUMOCOCCAL VACCINES FROM ANOTHER GENOMIC PERSPECTIVE (ID 399)
- Frank Esteban Mona, Colombia
- Frank Esteban Mona, Colombia
- Andrés Felipe Castro, Colombia
- Diego Andrés Sánchez, Colombia
- Yully Andréa Betancur, Colombia
- Alejandro Gómez, Germany
- Sven Hammerschmidt, Germany
- Gustavo Adolfo Gámez, Colombia
SEROTYPE-SPECIFIC URINE ANTIGEN DETECTION ENHANCES THE IDENTIFICATION OF NON-BACTEREMIC PNEUMOCOCCAL COMMUNITY ACQUIRED PNEUMONIA IN HOSPITALIZED CANADIAN ADULTS (ID 615)
- Jason J. LeBlanc, Canada
- Jason J. LeBlanc, Canada
- May ElSherif, Canada
- Lingyun Ye, Canada
- Donna MacKinnon-Cameron, Canada
- Ardith Ambrose, Canada
- Todd F. Hatchette, Canada
- Amanda L. Lang, Canada
- Hayley D. Gillis, Canada
- Irene Martin, Canada
- Walter Demczuk, Canada
- Craig LaFerriere, Canada
- Melissa K. Andrew, Canada
- Guy Boivin, Canada
- William Bowie, Canada
- Karen Green, Canada
- Jennie Johnstone, Canada
- Mark Loeb, Canada
- Anne E. McCarthy, Canada
- Allison McGeer, Canada
- Makeda Semret, Canada
- Sylvie Trottier, Canada
- Louis Valiquette, Canada
- Duncan Webster, Canada
- Shelly A. McNeil, Canada
GENOME-BASED APPROACH TO CHARACTERISE PNEUMOCOCCAL CARRIAGE DYNAMICS POST PCV-13 IMPLEMENTATION IN AN AFRICAN BIRTH COHORT: THE DRAKENSTEIN CHILD HEALTH STUDY (ID 929)
- Felix Dube, South Africa
- Felix Dube, South Africa
- Stephanie Lo, United Kingdom
- Chrispin Chaguza, United Kingdom
- Regina Esinam Abotsi, South Africa
- Rethabile Mokupi,
- Wendy S. Blose, South Africa
- Heather Zar,
- Mark Nicol, South Africa
MULTIPLEX PCR STANDARDIZATION FOR GENOTYPING OF STREPTOCOCCUS PNEUMONIAE IN CEREBROSPINAL FLUID (CSF) (ID 984)
- Mariana B. Souza, Brazil
- Mariana B. Souza, Brazil
- Maria Cecília C. Novela,
- Delma A. Molinari,
- Daniela R. Colpas,
- Andréia M. Carmo,
- Ivana B. Campos, Brazil
CLINICAL CHARACTERISTICS OF ADULT PNEUMONIA CASES IN THE ERA OF CHILDHOOD PCV13 VACCINATION IN MONGOLIA, 2015-2018 (ID 502)
- Claire Von Mollendorf, Australia
- Claire Von Mollendorf, Australia
- Mukhchuluun Ulziibayar, Mongolia
- Bujinlkham Suuri, Mongolia
- Dashtseren Luvsantseren, Mongolia
- Dorj Narangerel, Mongolia
- Rohini Beavon, United Kingdom
- Bradford D. Gessner, United States of America
- Tuya Mungan, Mongolia
- Kim E. Mulholland, Australia
HIGH-THROUGHPUT NANOFLUIDIC REAL-TIME PCR ASSAY TO EVALUATE SEROTYPE-SPECIFIC STREPTOCOCCUS PNEUMONIAE COLONIZATION IN THE NASOPHARYNX OF HEALTHY SOUTH AFRICAN CHILDREN. (ID 593)
- Sarah L. Downs, South Africa
- Sarah L. Downs, South Africa
- Shabir A. Madhi, South Africa
- Marta C. Nunes, South Africa
- Lara Van der Merwe, South Africa
- Courtney P. Olwagen, South Africa
NANOFLUIDIC REAL-TIME PCR TO DISCRIMINATE PNEUMOCOCCAL CONJUGATE VACCINE (PCV) ASSOCIATED SEROGROUP 6 TO INDIVIDUAL SEROTYPES. (ID 599)
- Sarah L. Downs, South Africa
- Sarah L. Downs, South Africa
- Shabir A. Madhi, South Africa
- Marta C. Nunes, South Africa
- Lara Van der Merwe, South Africa
- Courtney P. Olwagen, South Africa
INSIGHTS INTO PNEUMOCOCCAL PNEUMONIA USING LUNG ASPIRATES AND NASOPHARYNGEAL SWABS COLLECTED FROM PNEUMONIA PATIENTS IN THE GAMBIA (ID 668)
- Eileen M. Dunne, United States of America
- Eileen M. Dunne, United States of America
- Yinglei Hua, Australia
- Rasheed Salaudeen, Gambia
- Ilias Hossain, Gambia
- Malick Ndiaye, Gambia
- Belinda D. Ortika, Australia
- Kim E. Mulholland, Australia
- Jason Hinds, United Kingdom
- Sam Manna, Australia
- Grant Mackenzie, Gambia
- Catherine Satzke, Australia
A MOLECULAR ASSAY WITH INCREASED SENSITIVITY FOR IDENTIFYING COMMON BACTERIAL CAUSES OF PLEURAL EMPYEMA IN CHILDREN. (ID 732)
- Jonathan Jacobson, Australia
- Jonathan Jacobson, Australia
- Joshua Osowicki, Australia
- Loraine Fabri, Belgium
- Shivanthan Shanthikumar, Australia
- Eileen M. Dunne, United States of America
- Anna-Maria Costa, Australia
- Warwick Teague, Australia
- Jim Buttery, Australia
- Andrew Daley, Australia
- Andrew Steer, Australia
- Sarath Ranganathan, Australia
- Catherine Satzke, Australia
IMPACT OF MICROBIOLOGICAL WORKUP ON THE INCIDENCE OF HOSPITALIZATIONS DUE TO PNEUMOCOCCAL PNEUMONIA (ID 809)
- Julio Ramirez, United States of America
- Julio Ramirez, United States of America
- Stephen Furmanek, United States of America
- Senen Pena, United States of America
- Ruth Carrico, United States of America
- William Mattingly, United States of America
- Ronika Alexander, United States of America
- Kimbal D. Ford, United States of America
- Sharon L. Gray, United States of America
- Luis Jodar, United States of America
- Raul Isturiz, United States of America
- The Louisville Pneumonia Study Group, United States of America
HIGH INCIDENCE OF VIRAL CO-INFECTION DETECTION IN INDIAN CHILDREN UNDER 5 YEARS WITH PNEUMONIA - EVIDENCE FROM MULTIPLEX MOLECULAR ASSAY (ID 1136)
- Shincy M R, India
- Shincy M R, India
- KL Ravikumar,
Abstract
Background
Understanding the role of viral co-infection in defining the outcome of paediatric pneumonia will provide novel treatment and prevention strategies, improving patient’s outcome. The objective of this study was to determine the prevalence of viral infection in children with pneumonia
Methods
128 Nasopharyngeal swabs of children with clinically proven pneumonia collected from a tertiary care hospital in a duration of 1 year were tested. Multiplex real-time PCR assay- FTIyo Respiratory pathogens 33 test kit (Fast-Track Diagnosis, Luxembourg) was used for analysis.
Results
105/128(82%) of children included in the study tested positive for at least one pathogen: bacterial or viral. Viral infection was detected in 55/128(42%) of samples. Pure viral infection was present in 12%(7/55) of cases. Single viral co-infection accounted for 67%(37/55) of cases. Co-infection with duel virus was detected in 10 patients(18%) and triple infection in 1.8% (1/55).
The most common viruses identified were Adenovirus(47%), Bocavirus(32%), Rhinovirus (21%), Human Parainfluenza Viruses HPIV3(12%), HPIV4(3.63%) and Respiratory syncytial viruses A and B(3.63%) respectively
Conclusions
The study highlights the importance of evaluating viral co-infection in the context of pneumonia in children. It is an ideal approach for better understanding of the carriage with multiple viral agents and its relationship with the bacterial pathogen.
DEVELOPMENT AND VALIDATION OF A MACHINE LEARNING PREDICTION MODEL FOR PNEUMONIA MORTALITY IN CHILDREN AGED UNDER FIVE YEARS IN RURAL GAMBIA (ID 126)
- Alexander Jarde, Gambia
- Alexander Jarde, Gambia
- David Jeffries, Gambia
- Grant Mackenzie, Gambia
Abstract
Background
Pneumonia accounts for many deaths in children aged under 5 years in developing countries. A reliable and generalizable tool to predict mortality and thus assess the severity of pneumonia would aid patient management.
Methods
We used a dataset of 11,012 children admitted with clinical pneumonia to develop a model to predict mortality. Using a High Performance Computing platform, we generated multiple models for all possible feature combinations, applying support vector machine, neural networks, random forests and logistic regression to 2/3 of the dataset with repeated cross-validation (5 repetitions, 10 folds). We chose the final model based on its performance and on the number of and measurement reliability of the included features to increase generalizability. In the validation stage, we applied the selected model to the held-out dataset to test its performance on unseen cases.
Results
Not only did the selected model have good sensitivity and specificity (both >80%) on the training set, but more importantly, it had promising performance when applied to the test set.
Conclusions
Our predictive model performed well not only in cross-validated data, but also in our test dataset, increasing our confidence in its generalizability.
PNEUMOTAC: A HIGH-THROUGHPUT PLATFORM FOR SEROTYPING STREPTOCOCCUS PNEUMONIAE (ID 242)
- Jorge E. Vidal, United States of America
- Jorge E. Vidal, United States of America
- Fuminori Sakai, United States of America
- Srinivasan Velusamy, United States of America
- Mignon Du Plessis, South Africa
- Sibusisiwe Zulu, South Africa
- Anne Von Gottberg, South Africa
- Lesley McGee, United States of America
Abstract
Background
Sensitive, high-throughput platforms, for pneumococcal serotyping are essential for vaccine effectiveness studies. We developed PneumoTAC, a TaqMan array card (TAC) platform for the rapid identification of most S. pneumoniae serotypes.
Methods
PneumoTAC reactions underwent a thorough analytical validation including studies of limit of detection (LOD) and sensitivity. The performance of PneumoTAC was investigated on blinded pneumococcal isolates (N=131), and in N=84 clinical specimens from pneumococcal disease episodes [cerebrospinal fluid samples (CSF, N=71), and blood cultures (N=13)].
Results
The LOD of assays in PneumoTAC spanned between 2 and 21.4 genome equivalents. A sensitivity of 100%, and 98.8%, was achieved for S. pneumoniae species confirmation using DNA from isolates, and clinical specimens, respectively. The sensitivity for the detection of the serotypes/serogroups was 94% using DNA from isolates. In clinical specimens PneumoTAC detected a serotype in N=80 (96.3%) of samples, whereas a multiplex (m)PCR approach detected a serotype in N=55 (66%) of specimens. The correlation between the serotype obtained by mPCR and that obtained by PneumoTAC was 96.3% (N=53).
Conclusions
We developed PneumoTAC a multi-target platform with increased serotype coverage, compared to other TAC assays and multiplex PCR approaches, to identify 94 S. pneumoniae serotypes/serogroups in a single run.
ASSOCIATION BETWEEN C-REACTIVE PROTEIN LEVEL AND A RADIOLOGICAL END POINT CONSOLIDATION PNEUMONIA AMONG HOSPITALISED CHILDREN WITH SUSPECTED PNEUMONIA IN NEPAL (ID 618)
- Bibek Khadka, Nepal
- Bibek Khadka, Nepal
- Animesh Khulal, Nepal
- Sunaina Gurung, Nepal
- Meeru Gurung, Nepal
- Stephen Thorson, Nepal
- Shrijana Shrestha, Nepal
- Sanjeev M. Bijukchhe, Nepal
- Puja Amatya, Nepal
- Michael J. Carter, United Kingdom
- Matthew Smedley, United Kingdom
- Sarah Kelly, United Kingdom
- Kate M. Park, United Kingdom
- Merryn Voysey, United Kingdom
- David Murdoch, New Zealand
- Ganesh Shah, Nepal
- Maria Deloria Knoll, United States of America
- Dominic Kelly, United Kingdom
- Andrew J. Pollard, United Kingdom
Abstract
Background
S. pneumoniae is one of the most common causes of paediatric bacterial pneumonia. In low-income countries such as Nepal, CRP level and blood culture can be useful in diagnosis assessment. We assessed the association between CRP/blood culture, and pneumonia with end-point consolidation.
Methods
We included children less than 5 years of age admitted with suspected pneumonia to Patan Hospital in 2018 and 2019, whose chest xray, CRP level and blood culture were done. CRP levels >40 mg/dl were considered elevated.
Results
There was a significant difference (p<0.001) in CRP levels between EPC-pneumonia and non-EPC pneumonia cases with a median (IQR) CRP of 46.2 (16, 215) in 141 EPC-pneumonia cases and a median (IQR) CRP of 13 (4, 35) in non-EPC pneumonia cases. The sensitivity and specificity of CRP >40mg/dl to detect EPC pneumonia were 50% and 84% respectively. The area under the ROC curve was 0.727 indicating good discrimination between EPC-pneumonia and non-EPC pneumonia. Among the EPC-pneumonia cases, 62% had elevated CRP and 3.5% had S. pneumoniae positive blood cultures.
Conclusions
There was a significant association between CRP and EPC pneumonia. Blood culture had low sensitivity to detect bacterial pneumonia, nevertheless, CRP may be a useful tool in diagnosis of bacterial pneumonia.
MENINGOCOCCAL AND PNEUMOCOCCAL TYPING METHODS OF LOOP-MEDIATED ISOTHERMAL AMPLIFICATION (ID 641)
- Mitsuko Seki, Japan
- Mitsuko Seki, Japan
- Dong W. Kim, Korea, Republic of
- Paul Kilgore, Korea, Republic of
- Bing Chang, Japan
- Eun Jin Kim, Korea, Republic of
- Chika Takano, Japan
- Satoshi Hayakawa, Japan
- Tomonori Hoshino, Japan
Abstract
Background
We established meningococcal and pneumococcal typing methods of loop-mediated isothermal amplification (LAMP) and assessed their suitability for detecting meningococcal and pneumococcal types in cerebrospinal fluid (CSF).
Methods
The LAMP assays were developed to identify six major serogroups (A, B, C, X, Y, and W-135) of meningococcus and the 13 capsular types that are included in PCV13 (1, 3, 4, 5, 6A, 6B, 7F, 9V, 14, 18C, 19A, 19F, and 23F). We evaluated test reactivity, specificity, sensitivity, and compared the results between established LAMP and conventional PCR assays. To evaluate clinical performance of these assays, randomly selected CSF from children with suspected meningitis collected between 1998 and 2002 in Vietnam, China and Korea.
Results
The specificity of the primer sets was confirmed, and the assays could detect ten to 100 copies of purified DNA. The 31 meningococcal LAMP positive specimens were analyzed by serogrouping LAMP. The LAMP assay identified serogroups of 29 of the Neisseria meningitidis in CSF. The detection limits of the pneumococcal serotype specific LAMP assay were comparable when using DNA-spiked CSF, as compared to using purified DNA as the template.
Conclusions
Highly accurate LAMP assays have been developed for meningococcal and pneumococcal typing for globally use of surveillance and vaccine effectiveness studies.
THE EFFECT OF ANTIBIOTIC USE ON NASOPHARYNGEAL CARRIAGE OF S. PNEUMONIAE IN A SURVEILLANCE STUDY OF HOSPITALISED CHILDHOOD PNEUMONIA AND MALNUTRITION IN DILI, TIMOR-LESTE (ID 1029)
- Nevio D. Sarmento, Timor-Leste
- Nevio D. Sarmento, Timor-Leste
- Virginia Da Conceicao,
- Eloise Price,
- Paulino Da Silva,
- Katrina Lawrence,
- Heidi Smith-Vaughan, Australia
- Nicholas Fancourt, Timor-Leste
Abstract
Background
Hospital-based surveillance of pneumonia is an emerging method for monitoring pneumococcal conjugate vaccine (PCV) effectiveness, especially in low resource settings like Timor-Leste. We assessed the impact of antibiotic use on nasopharyngeal carriage of S. pneumoniae (SPN).
Methods
Pulmaun Saudavel is an ongoing pre-PCV surveillance study of children 1-59 months hospitalised with pneumonia and/or malnutrition. Nasopharyngeal swabs were collected as soon as practical after admission and cultured. Antibiotic exposure was classified as prehospital (by parent report) and in-hospital (from the medical record).
Results
Of 160 cases enrolled between September 2019 and January 2020, 43 (26%) had NP carriage of SPN. Carriage was higher among cases without any antibiotic exposure (15/27, 56%), compared to those with any antibiotic exposure (28/133, 21%; p<0.01). While there was no difference in carriage between those with and without prehospital exposure (24% vs 30%, p=0.39), those with swab collected <12hrs after in-hospital exposure had higher carriage compared to those with swab collected >12hrs (50% vs 19%, p=0.034). This remained significant in a logistic regression adjusted for age and sex (p=0.047).
Conclusions
Antibiotic exposure is associated with reduced NP carriage of SPN. Hospital-based studies should attempt to collect NP swabs within 12 hours of first antibiotic dose.
FINDING A SURFACE BIORECEPTOR FOR IDENTIFICATION OF STREPTOCOCCUS PNEUMONIAE. (ID 1081)
- José de Jesús Olivares-Trejo, Mexico
- José de Jesús Olivares-Trejo, Mexico
- Dan Israel Zavala-Vargas,
- Edgar Augusto Ortiz-Benítez,
- Juan Mosqueda,
Abstract
Background
Streptococcus pneumoniae is the principal etiological agent of otitis and meningitis in children around the world. In the last years a strain of this pathogen resistant to optochin has been detected. Therefore, there is a possibility to find false negatives in the current methods used to identify this pathogen. Then it is necessary to develop new identification methods.
Methods
Here, a strategy is presented to select a molecule that binds to S. pneumoniae surface, which would allow the specific identification of this pathogen. This strategy consists on using short single-stranded DNA (aptamer).
Results
The selected aptamer was able to bind the S. pneumoniae surface, but not to other bacteria such as: Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus pyogenes and Pseudomonas aeruginosa, with which can be confused during the identification processes. The aptamer selected was cloned in a plasmid vector and sequenced to know the sequence and to model the 3D structure. The aptamer was coupled to gold nanoparticles to obtain a bio-conjugated molecule and this structure also bound the S. pneumoniae surface indicating that the structure was not altered by the nanoparticles.
Conclusions
This aptamer is proposed as a bioreceptor to design a biosensor for the easy, fast and specific identification of S. pneumoniae.
TaqMan Array Card (TAC) for Identification of Pneumococcal Serotypes and Antibiotic Resistance Features (ID 481)
- Srinivasan Velusamy, United States of America
- Srinivasan Velusamy, United States of America
- Theresa Tran, United States of America
- Bernard Beall, United States of America
- Lesley McGee, United States of America
Abstract
Background
Identification of capsular serotype and antibiotic resistance (AR) profiles are both important strain features for pneumococcal disease surveillance. Our aim was to expand and validate a multiplex TAC (SPN_TAC) microfluidic platform to identify >90 serotypes, key AR and pilus genes on a single card.
Methods
Card included 75 assays designed to accurately identify 57 serotypes, 18 small serogroups, penicillin susceptibility, 2 pili genes and lytA and piaB positive controls for species confirmation. Probes were labelled with two fluorophores, FAM or ABY to detect 2 targets in a single reaction. LOD and specificity was validated with varying DNA concentrations of well characterized S. pneumoniae isolates from CDC’s ABCs collection.
Results
SPN_TAC could accurately identify 57 individual serotypes and 18 small serogroups, penicillin susceptibility, resistance to erythromycin, clindamycin, tetracycline and chloramphenicol and 2 pilus genes. Each assay had high sensitivity of detection (<10 copies/reaction) equivalent to CDC’s multiplex real-time PCR assays.
Conclusions
We developed and validated a highly sensitive multiplex method to identify pneumococcal serotypes/serogroups, key AR determinants and pili genes on a single TAC. The application of TAC for strain characterization of pneumococci allows for simple, fast, high-throughput testing that is suitable for surveillance and outbreak studies for both isolates and sterile-site clinical specimens.
COMMUNITY USE OF DIGITAL AUSCULTATION TO IMPROVE DIAGNOSIS OF CHILDHOOD PNEUMONIA IN SYLHET, BANGLADESH (ID 529)
- Salahuddin Ahmed, Bangladesh
- Salahuddin Ahmed, Bangladesh
- Eric D McCollum, United States of America
- Harish Nair, United Kingdom
- Steve Cunningham, United Kingdom
- Abdullah H. Baqui, United States of America
Abstract
Background
The WHO IMCI algorithm for childhood pneumonia diagnosis has high sensitivity but low specificity. This study aims to evaluate whether the use of automated lung sound classification through digital auscultation may improve the accuracy of pneumonia diagnosis in first-level facilities.
Methods
In a cross-sectional design, Community Health Workers (CHW) record lung sounds using a novel digital stethoscope (Smartscope) of 2426 under-5 children with possible pneumonia at first-level facilities in Bangladesh. A standardised paediatric listening panel is classifying the recorded sounds. A mobile app containing the Smartscope analysis system is also classifying the sounds and comparing with the reference paediatric panel’s classification.
Results
As of 31 December 2019, 1957 children screened, 1070 eligible cases identified and 1029 enrolled (32.67% had IMCI pneumonia). The results of the data collected during the first six months will be presented. These results will describe CHWs ability to record quality lung sounds and agreement between human and machine interpretation.
Conclusions
Auscultation and correct interpretation of lung sounds are often not feasible in the first-level facilities. Incorporation of the auto-classification of the Smartscope recorded lung sounds within the current IMCI pneumonia diagnostic algorithm may improve the accuracy of the diagnosis of childhood pneumonia at first-level facilities in LMICs.
CLINICAL FEATURES OF PNEUMONIA IN CHILDREN WITH AND WITHOUT SEVERE ACUTE MALNUTRITION (SAM): PRELIMINARY FINDINGS FROM A SURVEILLANCE PROJECT AT THE DILI NATIONAL HOSPITAL, TIMOR-LESTE (ID 679)
- Nicholas Fancourt, Timor-Leste
- Nicholas Fancourt, Timor-Leste
- Nevio D. Sarmento, Timor-Leste
- Edson Matoso,
- Narcisio Soares,
- Paulino Da Silva,
- Eloise Price,
- Milena Dos Santos,
- Joshua Francis,
Abstract
Background
SAM is a significant risk factor for pneumonia, but pneumonia can be hard to diagnose in children with SAM. We aimed to assess differences in clinical findings of pneumonia cases with and without SAM.
Methods
Pulmaun Saudavel is an ongoing surveillance study of children aged 1-59 months hospitalised with pneumonia and/or SAM in Dili, Timor-Leste. Pneumonia was defined as cough or difficulty breathing and any one of: respiratory rate >50bpm, oxygen saturation <90%, lower chest wall indrawing (LCWI), or World Health Organization(WHO)-defined radiological pneumonia. SAM was defined according to WHO criteria.
Results
Of 160 children enrolled between September 2019 and January 2020, 103 (64%) had pneumonia, 27 (17%) SAM, and 30 (19%) both pneumonia and SAM. Two children died, both with pneumonia and SAM. Compared to pneumonia-only cases, children with pneumonia and SAM more frequently met enrolment criteria because of radiological pneumonia (54% vs 42%) and less frequently for LCWI (48% vs 62%), and had more lethargy (43% vs 25%), and vomiting (17% vs 8%; all p>0.05). No differences were found for tachypnoea, hypoxemia, or auscultation findings.
Conclusions
Pneumonia is common in SAM. While LCWI is used in many pneumonia guidelines, it may not be as useful for children with SAM.
VACCINE-PREVENTABLE CLINICAL VERSUS RADIOLOGICALLY- OR ETIOLOGICALLY-CONFIRMED DISEASE IN PNEUMOCOCCAL CONJUGATE VACCINE EFFICACY TRIALS: A SYSTEMATIC LITERATURE REVIEW AND RE-ANALYSIS (ID 762)
- Bradford D. Gessner, United States of America
- Kaatje E. Bollaerts, Belgium
- Mark Fletcher, United States of America
- Jose A. Suaya, United States of America
- Germaine Hanquet, Belgium
- Marc Baay, Belgium
- Lindsay R. Grant, United States of America
- Christian Theilacker, Germany
- Thomas Verstraeten, Belgium
- Bradford D. Gessner, United States of America
Abstract
Background
While regulatory endpoints for pneumococcal conjugate vaccines (PCVs) routinely include radiologically- or vaccine serotype (VST) confirmed disease, public health decision-making benefits from consideration of all disease prevented regardless of diagnostic or etiological confirmation.
Methods
We followed PRISMA guidelines to perform a systematic literature review for Phase III/IV efficacy trials of PCVs from 1997 through 2019. We estimated study-specific vaccine-preventable disease incidence (VPDI) (control group minus intervention group incidences) for all-cause disease versus radiologically- and/or etiologically-confirmed outcomes. VPDI ratios between the broadest clinical and most narrowly defined outcomes were calculated [PROSPERO registration, CRD42019145268].
Results
Ten studies met the criteria. In children < 5 years, VPDI ratios ranged from 0.6 to 3.7 for otitis media (clinical versus VST etiologically-confirmed); from 1.3 to 1.8 for pneumonia (clinical versus radiologically-confirmed); 3.1 and 19.0 for pneumonia (clinical versus bacterial or VST); and 3.8 in one study of invasive pneumococcal disease (non-laboratory to laboratory-confirmed). In adults, VPDI ratios ranged from 2.2 to 2.9 for pneumonia (clinical to pneumococcal or VST).

Conclusions
Relying on only radiologically-confirmed or etiologically-confirmed outcomes substantially underestimated PCVs’ public health benefits. Broader clinical outcomes should be considered by vaccine technical committees when making decisions for pediatric and adult PCV use.
NASOPHARYNGEAL PNEUMOCOCCAL DENSITY IS ASSOCIATED WITH SEVERE PNEUMONIA IN YOUNG CHILDREN IN LAO PDR (ID 856)
- Fiona M. Russell, Australia
- Olivia J. Carr, Australia
- Keoudomphone Vilivong, Laos
- Mimee Laddaphone, Laos
- Eileen M. Dunne, United States of America
- Jana Y. Lai, Australia
- Jocelyn Chan,
- Malisa Vongsakid, Laos
- Chanthaphone Siladeth, Laos
- Belinda D. Ortika, Australia
- Mayfong Mayxay, United Kingdom
- Paul N. Newton, United Kingdom
- Lien Anh Ha H. Do, Australia
- Kim E. Mulholland, Australia
- Audrey Dubot-Pérès, Laos
- Catherine Satzke, Australia
- David A. Dance, Laos
- Fiona M. Russell, Australia
Abstract
Background
Pneumococcal nasopharyngeal colonisation density >6.9 log10 copies/mL is associated with primary endpoint pneumonia, very severe pneumonia and hypoxic pneumonia. Few studies have explored the association between pneumococcal density and severe pneumonia. We determined the association between nasopharyngeal pneumococcal density and children with severe pneumonia in Laos.
Methods
A prospective observational study was conducted at Mahosot Hospital. Children <5 years of age admitted with ARI were recruited (2014 to mid-2018). Clinical and demographic data were collected alongside with nasopharyngeal swabs. Severe pneumonia was classified according to the WHO 2013 definition. Pneumococci were detected and quantified by lytA qPCR. A logistic regression model deterimined the association between pneumococcal density and severe pneumonia, after adjusting for potential confounders.
Results
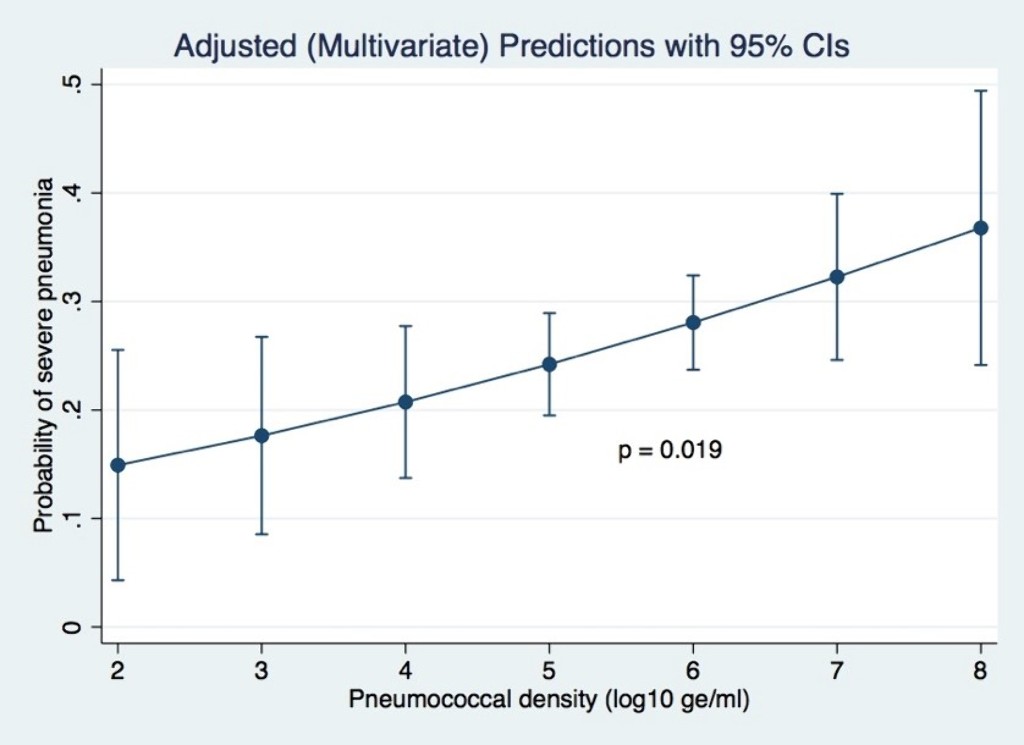
Of 1,289 participants enrolled, 32.2% had severe pneumonia. After adjusting for potential confounders (age, ethnicity, residential location, living with children <5 years, exposure to cigarette smoke, monthly income, PCV13 vaccination status and co-detection of RSV), pneumococcal density was positively associated with severe pneumonia (adjusted odds ratio 1.4; 95% CI 1.1–1.8; p=0.019).
Conclusions
Pneumococcal carriage density is associated with the probability of severe pneumonia in children in this setting.
DETECTION OF STREPTOCOCCUS PNEUMONIAE (SP) NASOPHARYNGEAL CARRIAGE (NPC) BY PCR ON DNA ISOLATED FROM PEDIATRIC NP SWABS PREVIOUSLY DETERMINED TO BE SP-NPC NEGATIVE BY CULTURE (ID 916)
- Michelle H. Davis,
- Michelle H. Davis,
- Amy Tunali, United States of America
- Stepy Thomas,
- Robert C. Jerris,
- Monica Farley, United States of America
- Sarah W. Satola, United States of America
Abstract
Background
Monitoring Streptococcus pneumoniae nasopharyngeal carriage (SP-NPC) and SP serotype distribution is critical to evaluate the impact and efficacy of pneumococcal vaccine programs. The gold standard for pneumococcal colonization and serotype distribution is culture, followed by serotyping with antisera. Detection of very low density SP colonization or concurrent colonization by multiple serotypes may be missed by culture. Real-Time PCR (RT-PCR) targeting SP genes lytA (major autolysin) and SP2020 (putative transcriptional regulator) has been identified as a powerful and sensitive molecular strategy for identification of SP.
Methods
NP swabs collected from children <5 years in an emergency department in Atlanta, GA in 2017 were broth-enriched, cultured for SP, and serotyped. RT-PCR targeting lytA and SP2020 was performed on all NP swabs negative for SP by culture. Subsequent RT-PCR targeting capsule polysaccharide-specific cpsA, associated with encapsulated SP strains, was performed on all ltyA and SP2020-positive/culture-negative NP swabs.
Results
622 children were enrolled; 182 (29.3%) had SP-NPC by culture. Of the 440 culture-negative, 59/440 (13.4%) were positive for lytA/SP2020 with 45/59 (76.2%) cspA+.
Conclusions
Molecular detection of SP from NP swabs increased carriage from 29.3% to 38.7%. 76.2% of SP identified by RT-PCR appear to be encapsulated and molecular serotyping is currently underway.
RAPID INFERENCE OF ANTIBIOTIC RESISTANCE AND SUSCEPTIBILITY BY GENOMIC NEIGHBOR TYPING (ID 1213)
- Karel Břinda, United States of America
- Karel Břinda, United States of America
- Alanna Callendrello,
- Kevin C. Ma,
- Derek MacFadden,
- Themoula Charalampous,
- Robyn S. Lee,
- Lauren Cowley,
- Crista Wadsworth,
- Yonatan Grad,
- Gregory Kucherov,
- Justin O’Grady,
- Michael Baym,
- William Hanage, United States of America
Abstract
Background
Surveillance of drug-resistant bacteria is essential for healthcare providers to deliver effective empiric antibiotic therapy. However, traditional molecular epidemiology does not typically occur on a timescale that could impact patient treatment and outcomes.
Methods
We developed a method called ‘genomic neighbor typing’ for inferring the phenotype of a bacterial sample by identifying its closest relatives in a database of genomes with metadata. We implemented this with rapid k-mer matching, which can be used on Oxford Nanopore MinION data and run in real time. We adopted the method for pneumococcus using a database of 616 genomes from a carriage study in Massachusetts children and five antibiotics. Finally, we evaluated the method using isolates and metagenomes from geographically distinct regions.
Results
We show that genomic neighbor typing can infer antibiotic resistance and susceptibility of S. pneumoniae isolates within five minutes of sequencing starting (sens/spec 91%/100%) and for clinical metagenomic sputum samples within four hours of sample collection (75%/100%). We also show how the method can be adopted for custom species and drugs.
Conclusions
Genomic neighbor typing has wide application to pathogen surveillance and may be used to greatly accelerate appropriate empirical antibiotic treatment.
PACBIO SMRT AMPLICON SEQUENCING FOR STRAIN BASED PNEUMOCOCCAL RESOLUTION (ID 1220)
- Oluwaseun Oyewole, Switzerland
- Oluwaseun Oyewole, Switzerland
- Markus Hilty, Switzerland
Abstract
Background
Current methods for detecting pneumococcal co-colonisation often rely on a culturing step. Here, we present an improved direct sample detection method for distinguishing pneumococci through single molecule real time (SMRT) sequencing of the ~1.4kb pneumococcal molecular marker, plyncr.
Methods
A set of nine mock communities from in-house strains with known plyncr signatures was created in varying ratios up to two orders of magnitude. A second set of 14 nasopharyngeal swab (NPS) samples (previously analyzed by plyNCR-RFLP) was blindly prepared. Plyncr-PCR and Sequel sequencing were performed. The R package, DADA2, was used to detect amplicon variants. RFLP signatures were identified with the online restrictionmapper tool.
Results
Within the first run set, DADA2 accurately deconvoluted 8 out of 9 communities (89% accuracy) including two deepest communities at ratios 1:89 and 1:99. Blinded analysis of the 14 NPS samples revealed 100% accuracy of dominant strain identification, of which in only 46%, minor strains were not detected.
Conclusions
SMRT sequencing of the plyncr marker allows for accurate identification of pneumococcal strains to 1% variant level. Further results from an ongoing carriage study in newborn Swiss infants up to 1 yr of age using this method may provide better understanding of the dynamics of pneumococcal acquisition and carriage.
SEROGROUP 35 RESPONSES IN QUELLUNG REACTION AND ANALYSIS USING WHOLE GENOME SEQUENCING OF NASOPHARYNGEAL AND MIDDLE EAR FLUID ISOLATES FROM CHILDREN IN ROCHESTER NY (ID 954)
- Naoko Fuji, United States of America
- Naoko Fuji, United States of America
- Ravinder Kaur, United States of America
- Joshua Mell,
- Rachel Ehrlich,
- Joshua Earl,
- Garth Ehrlich,
- Michael Pichichero, United States of America
Abstract
Background
For serogroup 35, definition of serotype 35A occurs if reactions are interpreted as negative using sera specific for 35B, 35C and 35F.
Methods
From 2015-2019, a random subset of pneumococci serogroup 35 nasopharyngeal and middle ear fluid isolates from Rochester NY children were analyzed using Quellung and whole-genome sequencing (WGS).
Results
23 isolates positive for serogroup 35 by Quellung analyzed by WGS showed that 18 strains had been identified as serotype 35A by Quelling that were 35B by WGS. The 18 strains were retested by Quellung and 14 were reconsidered as positive with serotype 35B typing sera. 4 isolates were repeatedly negative for 35B by Quellung. We analyzed the genetic features of these 4 atypical 35B isolates. One isolate had a mutation on wciB (Y to C). Analysis of noncapsular region identified one region showing low similarity to the typical 35B (35B in Quelling reaction and WGS). That region was about 35K bp, coding 31 proteins that showed less than 75% similarity with typical 35B strains in DNA.
Conclusions
Caution is essential during interpretation of Quellung reactions for serogroup 35. We identified 4 atypical strains of serotype 35B that may have differential capsule expression. Further analysis is ongoing.