OO018 - DEEP LEARNING OF TRANSCRIPTOMIC DYNAMICS TO PREDICT HIPPOCAMPAL ATROPHY (ID 675)
Abstract
Aims
Hippocampal atrophy occurs early in Alzheimer’s disease (AD). The peripheral blood transcriptome reveals changes in gene expression profiles. The understanding of how the alterations of gene expression relates to hippocampal atrophy is still limited. We leveraged a novel deep learning approach to analyze how transcriptomic changes relate to hippocampal atrophy.
Methods
We used imaging and transcriptomic ADNI data from 426 patients (CN=137, MCI=289), the small size AD group is not used to avoid unbalanced dataset (Table 1). We designed a multi-stage deep learning architecture to learn the hidden patterns of the transcriptomic data. The high-dimensional transcriptomic vector was reduced by K-means clustering and principal component analysis, which selected the representative genes. The generated gene expression vector, along with age and gender information, were then fed into the neural network aiming to predict hippocampal atrophy. The patterns hidden in the high dimensional feature vector were abstracted layer by layer in the network. The learning process is based on the cross entropy loss.
| Demographic | CN (n=137) | MC (n=289) |
| Age, Mean (SD) | 73.11(6.03) | 70.88(7.30) |
| Gender (M/F) | 64/73 | 153/136 |
Results
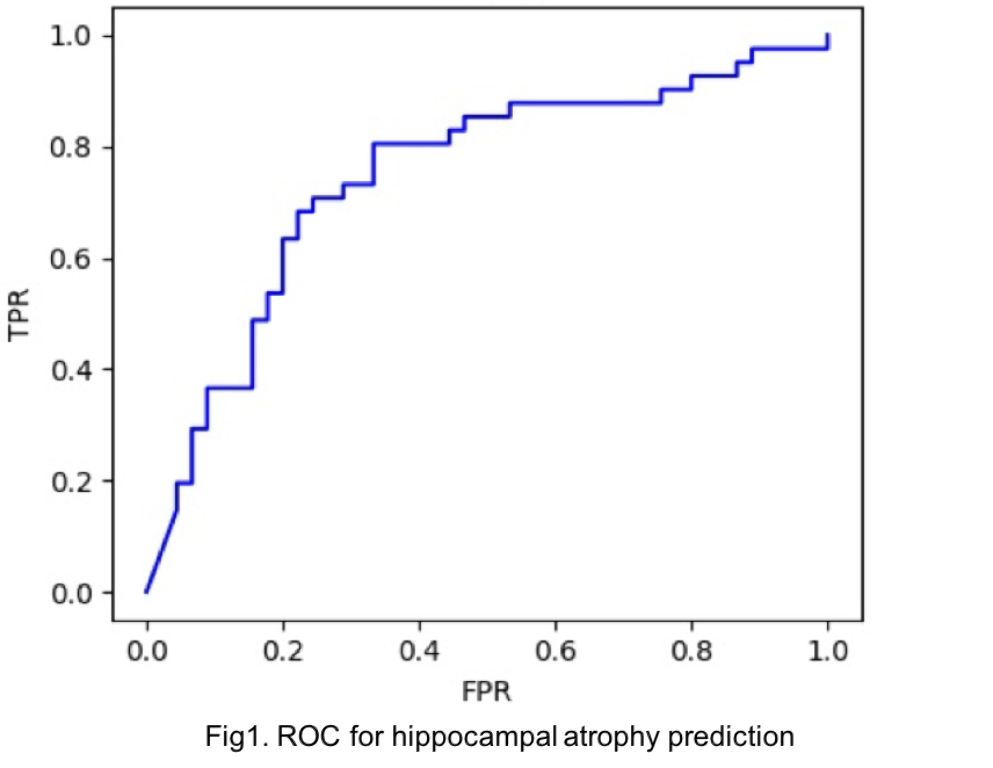
The dataset was split into 80% and 20% for training and testing purposes. The 49,386-dimension transcriptomic vector was reduced to 3,482-dimension. Using the 3,482 transcripts, our deep learning model achieved an accuracy of 73% and an area under the curve (AUC) of 74% in predicting hippocampal atrophy (Fig.1).
Conclusions
Our novel deep learning approach shows promising results and is expected to greatly advance the application of deep learning in AD research.











