Ignacio Hidalgo, Spain
Universidad Complutense de Madrid Computer Architecture and AutomationPresenter of 3 Presentations
PROFILED GLUCOSE FORECASTING USING GENETIC PROGRAMMING AND CLUSTERING
Abstract
Background and Aims
This research is motivated by the challenge of creating a general method to accurately predicting future glucose levels, so, an automated or manual system can decide when and how much insulin to inject in order to maintain glucose levels within a healthy range. It is imperative to avoid predictions that may trigger unnecessary treatments or, even worse, treatments that are harmful to the patient.
Methods
The proposed methodology is a three-step process: Data collection, preprocessing, and division, Data clustering and detection of a set of glucose profiles, and, Models training, creating models by evolutionary computation. CHAID (CHi-square Automatic Interaction Detection) is applied to recursively divide the data in relation to a target variable using multiple divisions between the different input variables: days of the week and time slots. A data augmentation algorithm hat generates synthetic glucose time series is used in training datasets to develop meaningful information and significantly enhance data quality. Next, models based on Genetic Programming (GP) are created using cross-validation. Then, the best model of 10 repetitions is selected by the Akaike Information Criterion (AIC).

Results
Data was collected from ten patients. The predictors used in the construction of the tree are the day of the week and the time slot. Significant differences were observed in the glucose profiles classified for each of those categories.
Conclusions
The accuracy of predictions with models created with GP is better for shorter time horizons and gradually gets worse as the time horizon increases from 30 to 240 minutes.
Thanks: RTI2018-095180-B-I00 and Fundación Eugenio Rodriguez Pascual
GLUCOSE FORECASTING WITH RANDOM GRAMMATICAL EVOLUTION
Abstract
Background and Aims
Bolus decision is a difficult task since patients need to estimate the number of carbohydrates they are going to ingest, take into account the past and future circumstances, know the past values of glucose, evaluate if the effect of previously injected insulin has already finished and any other relevant information. In this work, we present and compare a set of methodologies to automate the decision of the insulin bolus, which reduces the number of dangerous predictions.
Methods
We combine two different data enrichment techniques based on Markov chains with grammatical evolution engines to generate models of blood glucose, and univariate marginal distribution algorithms and bagging techniques to select the set of models to assemble. Modeling is solved as four symbolic regression problems by Random-Grammatical Evolution.
Results
We report the Clarke’s Error Grid Analysis.The best results are obtained with an ensemble model developed by bagging of 100 models. For a 30 minutes forecasting horizon, all the strategies have similar performance with almost no points in the D and E zones. For the 60, 90 and 120 minutes, the best models are produced by Random-GE and Bagging with both data augmentation techniques, obtaining a 95% of safe predictions,on average.
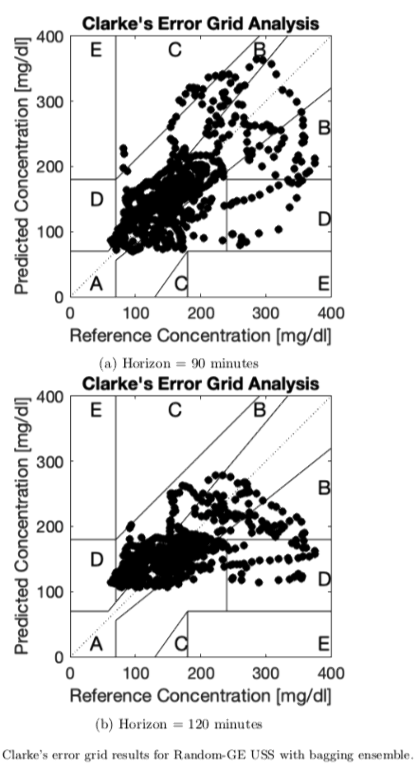
Conclusions
Results show how the methods improve results from pre-vious works. The proposal uses a simplified preprocessing process of the raw data, we use symbolic regression standard grammars and execution times are low. In future works, we should move a step forward in the forecasting horizon.
Thanks: RTI2018-095180-B-I00 and Fundación Eugenio Rodriguez Pascual
A WEB APPLICATION FOR THE IDENTIFICATION OF BLOOD GLUCOSE PATTERNS THROUGH CONTINUOUS GLUCOSE MONITORING AND DECISION TREES
Abstract
Background and Aims
This increased demand for CGM devices means an opportunity for data and computer scientists, who can contribute to the development of decision-making support systems based on the data collected from the devices. Our aim is to applied Machine Learning techniques to this data and find patterns that leads to advisory systems.
Methods
Using both the entered data and the blood glucose values collected by the device automatically, the application presented here uses decision trees to detect the patterns and entails a starting point in the creation of ensemble models with more predictive power, also based on decision trees. Furthermore, the methodology makes a segmentation of the data set in blocks, determined by the different meals done throughout the day, adding more information to the set of variables used to train the models.
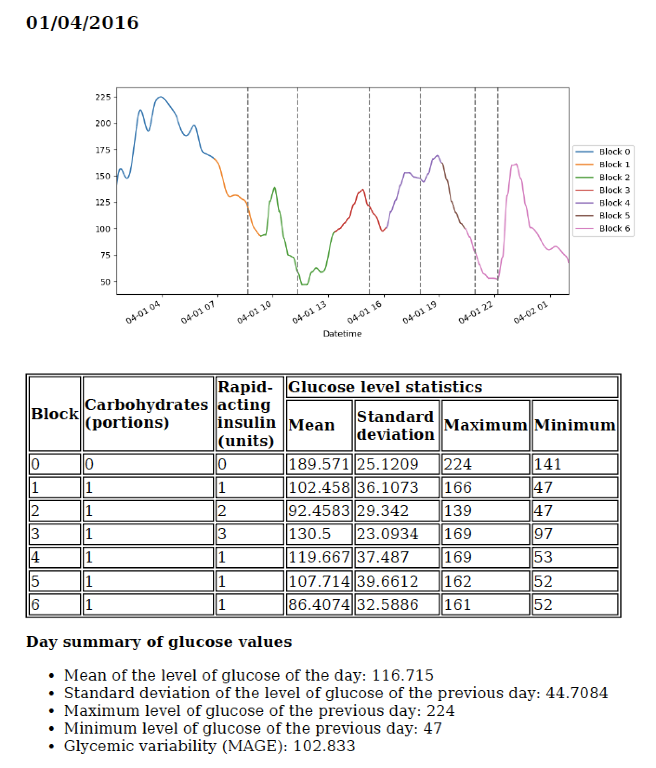
Results
The application developed in this project generates a report of the patient’s glucose patterns and provides a web application that allows the user to upload the data obtained from his device and download the report on his computer or smartphone.
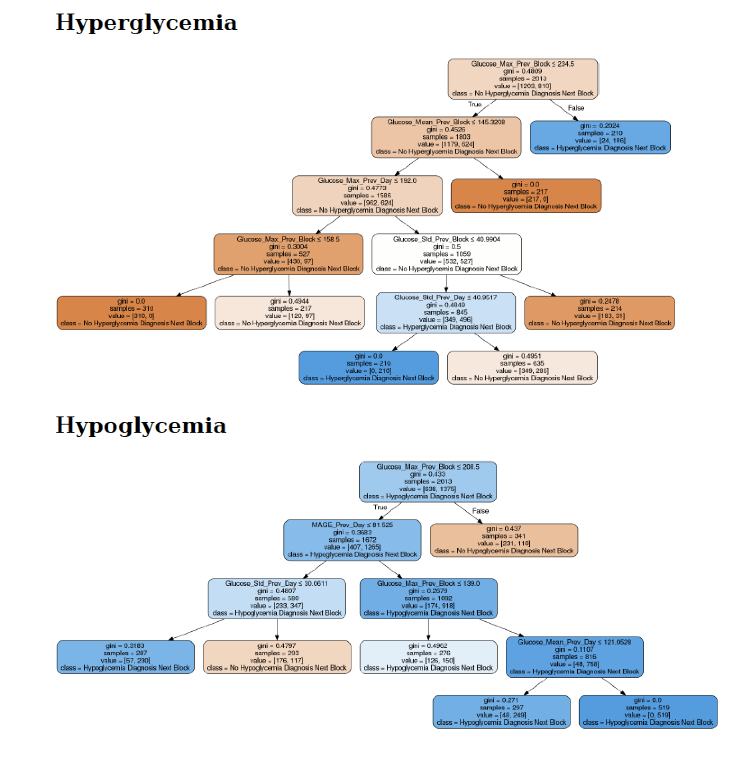
Conclusions
As a result, the application can discover repetitive patterns in the daily life of the patient, which can help him to take early preventive measures for risk situations in a period close to the next meal.
Thanks: RTI2018-095180-B-I00 and Fundación Eugenio Rodriguez Pascual