Abstract
Background and Aims
In recent years, neural network models (NNMs) have gained popularity for use in predicting blood glucose (BG) values. These models have the advantage of being data-driven and trained to model an individual's unique physiological characteristics. However, NNMs remain opaque and difficult to understand and interpret by human users. In this work, we address a key challenge in designing and training NNMs for BG prediction: ensuring NNMs conform to known physiological dynamics. We demonstrate how the standard learning protocol results in non-conformant models, and present a physiologically-motivated learning approach to ensure conformance is obtained without sacrificing accuracy.
Methods
Using a combination of mixed integer programming and local gradient search, maximum and minimum changes in predicted BG corresponding to increased insulin infusions are tested in three classes of NNMs: a generalized NNM structure, and two novel physiologically motivated structures. Model sensitivity of each input is computed and tested for conformance to "increased insulin results in decreased BG values". Models are trained using previously collected CGM and insulin pump data from a cohort of twenty-four subjects with T1D to predict 60min horizons.
Results
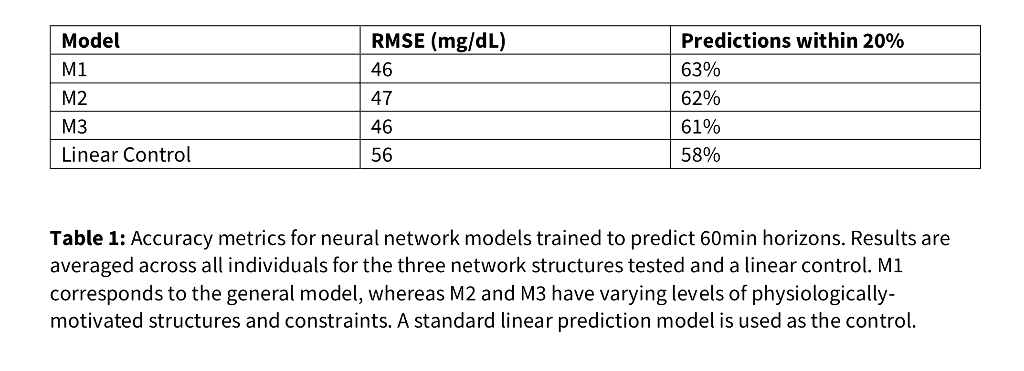
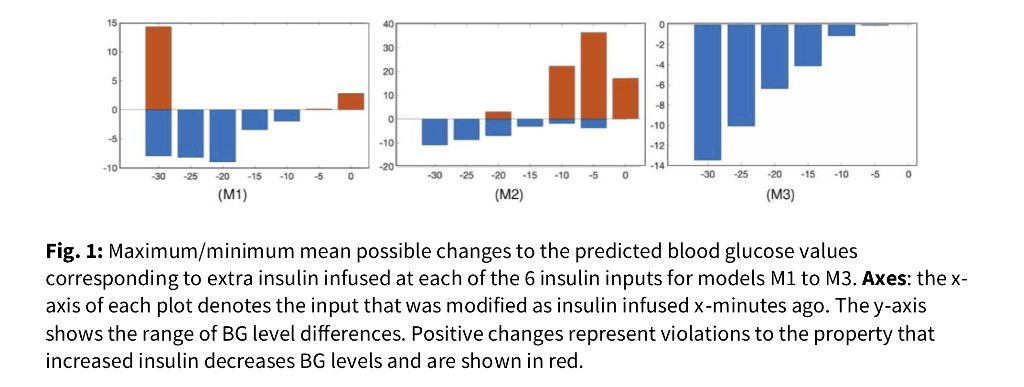
Accuracy, computed by root mean square error, remains consistent across all models [Table 1]. However, only the constrained, physiologically motivated model fully conforms to "increased insulin decreases BG" [Fig. 1].
Conclusions
A novel test for conformance of NNMs to known physiology is presented. We demonstrate how standard NNMs learned for BG prediction can fail to conform, and present a physiologically motivated design to learning which improves conformance while maintaining accuracy.